CAFA PI Data Exploration
The Challenge
This semester, I’ve been working with the BYU Bioinformatics Research Group on the CAFA PI challenge. The goal of CAFA PI is to predict which proteins perform some specific functions. The first major roadblock: no training data was provided.
So we divided into three groups:
- Data acquisition (we needed to get our hands on some training data),
- Data transformation (said training data needed to be formatted), and
- Data analytics.
The goal of this post is to explore the test data in order to inform our future acquisition / tranformation / analytics decisions.
[Insert cool meme here]
The Data
Targets
CAFA PI gives us protein sequences for two organisms:
- Pseudomonas aureginosa (this is the spelling the CAFA website gives, but it seems like the correct spelling is aeruginosa), a multidrug resistant bacteria that’s associated with, among other nasty things, hospital-acquired infections.
- Candida albicans, an opportunistic yeast that’s found in about half of all healthy adults but that will also not hesitate to turn on you as soon as your immune system goes down. Wikipedia says that somewhere between 2,800 and 11,200 people in the U.S. die from Candida infections every year.
The training data comes in FASTA format, so our first order of business is to turn the FASTAs into a tidy, rectangular format. Code to download and parse the data into CSVs can be found on our GitHub repo.
Let’s load the data in and take a quick look:
pseudomonas <- read_csv("data/cafa-pi/target.208963.csv")
pseudomonas$Organism <- "Pseudomonas aureginosa"
candida <- read_csv("data/cafa-pi/target.237561.csv")
candida$Organism <- "Candida albicans"
targets <- bind_rows(pseudomonas, candida)
targets %>% select(-Sequence) %>% head() %>% kable()| CAFA_ID | Taxon_ID | NCBI_Locus_Tag | Organism |
|---|---|---|---|
| T2089630000001 | 208963 | PA14_31220 | Pseudomonas aureginosa |
| T2089630000002 | 208963 | PA14_64790 | Pseudomonas aureginosa |
| T2089630000003 | 208963 | PA14_35250 | Pseudomonas aureginosa |
| T2089630000004 | 208963 | PA14_48450 | Pseudomonas aureginosa |
| T2089630000005 | 208963 | PA14_29300 | Pseudomonas aureginosa |
| T2089630000006 | 208963 | PA14_27470 | Pseudomonas aureginosa |
targets$Sequence %>% head()## [1] "MSSHHDYIIEITAQHDALKPFAPENGQPLRFKIGDAVIYTNEYGVQFRRCVTGFYRPSGLCGHYARGARYLLNSTSPWVPVAESSLRPDDSA"
## [2] "MNRDLRRALDDEPMRPFQWLAVAVCIVLNLIDGFDVLVMAFTASSVAAEWNLGGAEIGLLLSAGLFGMAAGSLFIAPWADRWGRRPLILACLALSGLGMLASALSQAAWQLALLRGLTGLGIGGILASSNVIASEYASRRWRGLAVSLQSTGYALGATLGGLLAVWLIGAWGWRSVFVFGAGLTLAVIPLVCLCLPESLDFLLARRPPRALQRVNALARRLGRAALAELPSPPGGGAEHGSRLARLLCVAQRRTTLLLWALFFLVMFGFYFIMSWTPKLLVAAGLSTAQGITGGTLLSIGGIFGAALLGGLAARFRLERVLALFMLLTAALLALFSLSAGLPGAALPLGLLIGLCANACVAGLYALAPSLYDASVRATGVGWGIGVGRGGAILSPLVAGLLLDDGWQPLSLYGAFAAVFVAAAAVLPLLGARRRERSPTLGDAA"
## [3] "MQKMNAPLQYRLDHADLALVLALVRGGSLARAAELLRVDVSTVFRAIRRLEKNLGKALFDKSRSGYLANQTARRLAQQAELAEQALEAARIGLELEREVLSGTVRLTCSDAVLHGLLLPALARFMPGYPALQLELATSNDFANLSRRDADIALRMTRTPPEHLVGRNLGEVRYVLCASERYLEGRDATEPAMLHWIAPDDFLPDHPSVVWRRRHYPGVLPAYRCNGVLAVSEMARAGLGLAAIPAYLLRDGDGLRVLDADLEGCASALWLLTRPDCRALRSVVALFDELGRHVRLGDE"
## [4] "MTRRHFLQRLGASAGLGAALTLGLEFGSPRGQAAAADHWHMPDEHLPQERVFLAYAASPSIWKDLAEDVNRSVALLARTIARYQPVTLLCRPTQEAAARRACGGANIDYLALPLDDIWVRDYGGCFVLNGEGEAGLVDFNFNGWGGKQQAGNDSRVAGVLSERLEVRYIASQLTGEGGGIEVDGRGTAILTESCWLNDNRNPGMDKGQVEAELKALLGLRKIVWLPGIRGRDITDAHVDFYARFVRPGVVIANLDNDPDSYDYAVTREHLEILRTATDADGRALQVHTLSPPLKPRRDYSRNNPDFAAGYINYLPINGAVIAPQFGDASADRHCGELLGRLYPGRDIVQLNIDPIAAGGGGIHCVTKHMPRA"
## [5] "MALYSAGVEYGIHCLLFLADEKGDSRESSVRTLAELQGVPPELLAKVFTRLAKAGLVAATEGVRGGFRLARPANEISVLDVVRAIDGDKSIFECREVRERCAIFEGNPPSWATRNTCSIHAVMLTAQKRMEEALAQQTILDLVRRVGRIAPPEFGEEVLRWMDASREGRGGSRD"
## [6] "MQIRADFDSGNIQVIDASDPRRIRLAIRPDLASQHFQWFHFKVEGMAPATEHRFTLVNAGQSAYSHAWSGYQAVASYDGERWFRVPSQYDADGLHFQLEPEESEVRFAYFKPYSRERHARLVERALGIEGVERLAVGTSVQGRDIELLRVRRHPDSHLKLWIIAQQHPGEHMAEWFMEGLIERLQRPDDTEMQRLLEKADLYLVPNMNPDGAFHGNLRTNAAGQDLNRAWLEPSAERSPEVWFVQQEMKHHGVDLFLDIHGDEEIPHVFAAGCEGNPGYTPRLERLEQRFREELMARGEFQIRHGYPRSAPGQANLALACNFVGQTYDCLAFTIEMPFKDHDDNPEPGTGWSGARSKRLGQDVLSTLAVLVDELR"The first thing that really stands out here is the huge difference in lengths of the proteins. Let’s look at how sequence length is distributed.
targets <- targets %>% mutate(seq_length=nchar(Sequence))
targets %>% select(seq_length) %>% summary()## seq_length
## Min. : 23.0
## 1st Qu.: 213.0
## Median : 353.0
## Mean : 434.6
## 3rd Qu.: 556.0
## Max. :5212.0targets %>% ggplot(aes(x=seq_length)) +
geom_histogram(aes(fill=Organism), bins=50) +
theme(legend.position="bottom") +
labs(x="Sequence Length", y="Count",
title="Sequence Lengths of CAFA PI Targets")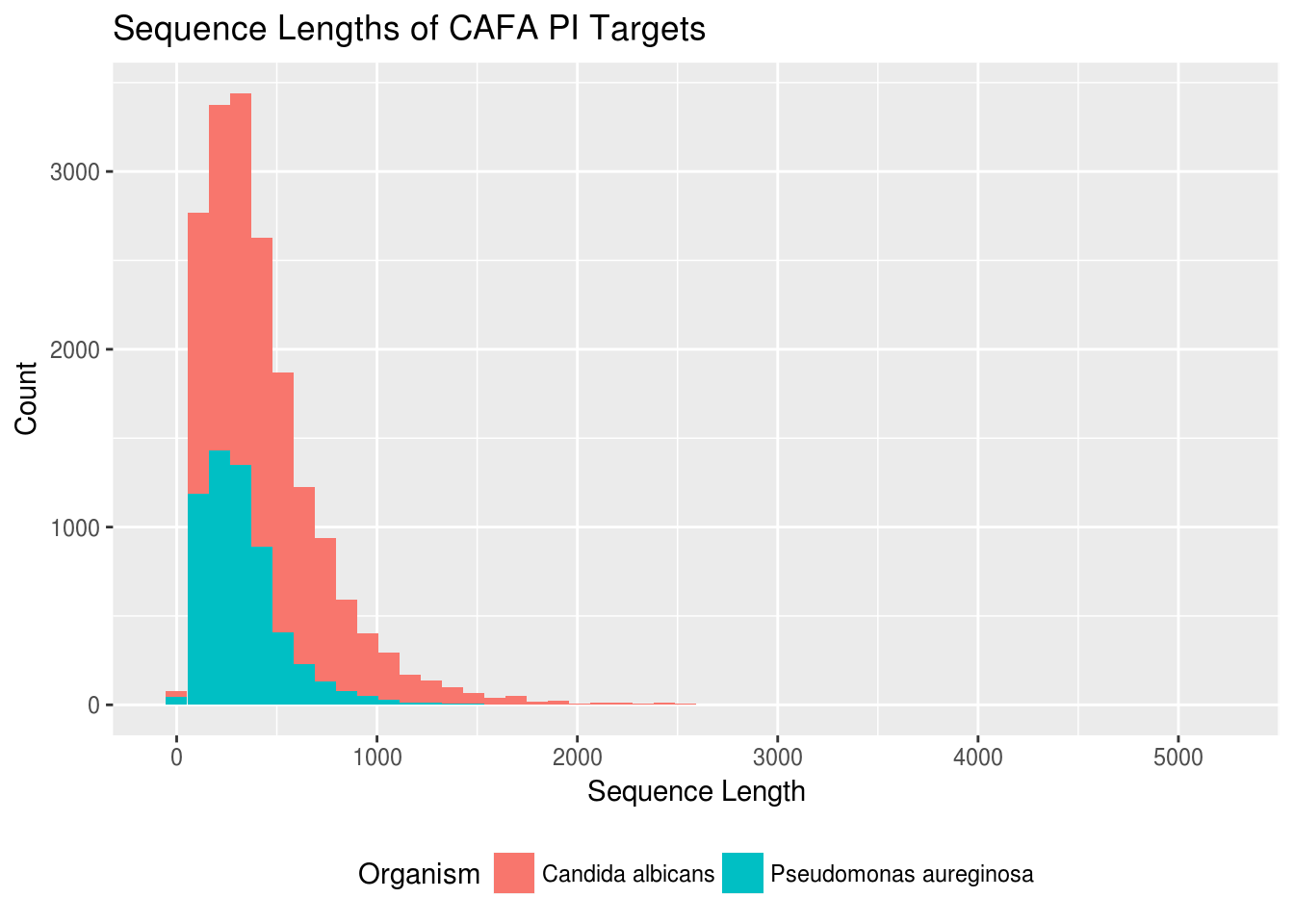
It looks like we have more yeast proteins than bacteria proteins and that on average the yeast proteins are longer. I don’t know a ton about the biology of proteins, but this makes intuitive sense to me—bacteria are prokaryotes, and yeast are eukaryotes, so I would expect yeast to have more and more complex proteins.
Now let’s take a look at that tail of super long proteins, just to see if it matches what we’d expect.
targets %>% filter(seq_length > 2500) %>%
ggplot(aes(x=seq_length)) +
geom_histogram(aes(fill=Organism), bins=1000) +
theme(legend.position="bottom") +
labs(x="Sequence Length", "Count",
title="The Tail")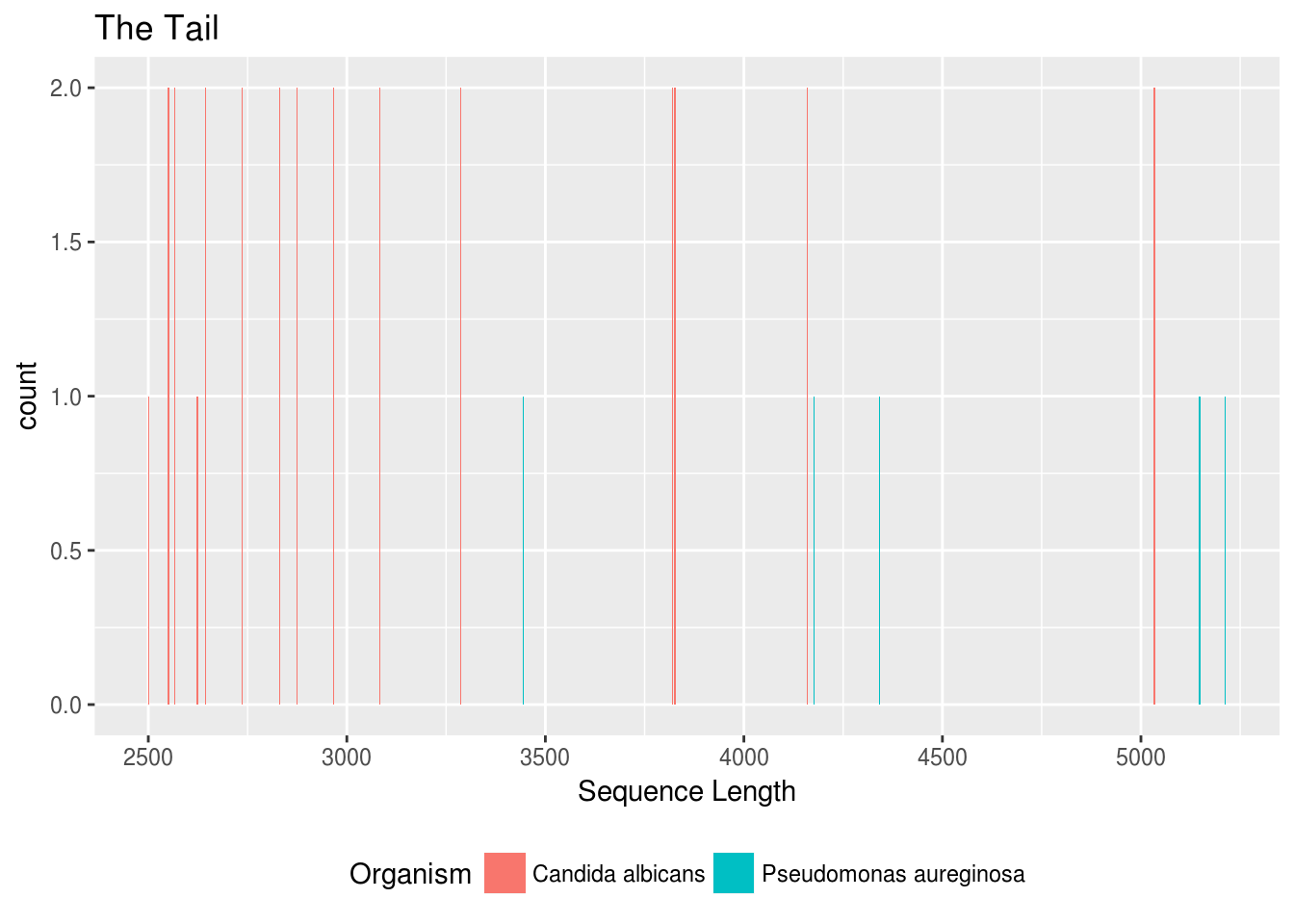
Two things here are a little interesting / unexpected to me:
- Our longest protein is from our bacteria, not our yeast. I’m not too freaked out about this, but we should take a look at that long bacteria protein just to make sure it’s OK.
- Our yeast proteins seem to almost exclusively come in pairs. It seems really unusual to me that all of these really long proteins would have a different protein with exactly the same length.
OK, let’s take a look, starting with the long bacterial proteins.
targets %>% arrange(desc(seq_length)) %>%
filter(Organism == "Pseudomonas aureginosa") %>%
select(NCBI_Locus_Tag, seq_length) %>%
head()## # A tibble: 6 x 2
## NCBI_Locus_Tag seq_length
## <chr> <int>
## 1 PA14_32790 5212
## 2 PA14_33610 5149
## 3 PA14_33280 4342
## 4 PA14_55400 4177
## 5 PA14_00510 3443
## 6 PA14_05390 2476A quick Googling of the NCBI Locus Tags shows that these six longest bacteria sequences all have transposon mutants (for example, PA14_32790). Not positive what that means in this context, but a transposon is a chunk of DNA that can jump into other places in DNA, so maybe these sequences are so long because extra DNA has jumped into them? Or maybe “transposon mutants” means something else. Note: Look to see if shorter sequences have transposon mutants.
Now let’s look to see about duplicates in our yeast.
targets %>% arrange(desc(seq_length)) %>%
filter(Organism == "Candida albicans") %>%
select(NCBI_Locus_Tag, seq_length, Sequence) %>%
# Trim sequences to the first 100 chars so they display nicely
mutate(Sequence=str_c(str_sub(Sequence, 1, 100), "...")) %>%
head()## # A tibble: 6 x 3
## NCBI_Locus_Tag seq_length Sequence
## <chr> <int> <chr>
## 1 C4_00970C_A 5035 MNNHITISFNQGEELYSTYKSFYTLKKLPNQLFKFNKSLSIDEILNQLSLLALDHTLPTYYCYKPIFLELVARWVNNTIELESQYNKSKSETRRISGSVI...
## 2 C4_00970C_B 5035 MNNHITISFNQGEELYSTYKSFYTLKKLPNQLFKFNKSLSIDEILNQLSLLALDHTLPTYYCYKPIFLELIARWVNNSIELESQYNKSKSETRKIPGSII...
## 3 C3_05210C_B 4161 MEESVTPLLSVNQLYDCIIGFLPFNENVPKFTQCEDVLTKFISNNNQDTLYLIKSIEEKTSRISNDLSDLDMGFKDLDTFVIIIKSKGELRNDIPLTQQL...
## 4 C3_05210C_A 4161 MEESVTPLLSVNQLYDCIIGFLPFNENVPKFTQCEDVLTKFISNNNQDTLYLIKSIEEKTSRISNDLSDLDMGFKDLDTFVIIIKSKGELRNDIPLTQQL...
## 5 C4_06130W_A 3826 MSSKEKPSKEDIRGQLSDIQKRLFSKKPSSGDTSGATTSASTNSNHRPVLKESNHHPQGKSSLLSGGVGSHEQERFADFLPPLPKGSGNASASLGPPAPT...
## 6 C4_06130W_B 3826 MSSKEKPSKEDIRGQLSDIQKRLFSKKPSSGDTSGATTSASTNSNHRPVLKESNHHPQGKSSLLSGGVGSHEQERFADFLPPLPKGSGNASASLGPPAPT...Looks like our identical lengths are from different alleles (slightly different copies of the same gene due to genetic variation). Now we know! It will be interesting to see whether our classifier predicts the same functions for two alleles of the same gene.
